DP12 Imputation using LeftJoin and Coalesce
Description
Another example of imputation of missing values but in this case using the information located in the same data table, for the same MID but in other periods. This was used on demographic values as for example race, DOB, that we can assume don’t change for the same individual. A table without missing values is created using dplyr::group_by. Then the original data is merged with dplyr::left_join(). Finally the replacing value is taken with coalesce.
Link to the Complete Script in Github
R Script - Inputation using LeftJoin and Coalesce
Initial data with missing values
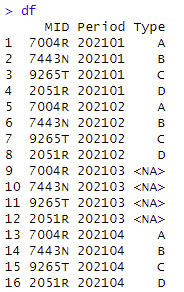
Code to create a new data frame by excluding the rows with missing (NA) values:
1
2
3
4
df %>% dplyr::arrange(Period) %>%
filter(!is.na(Type)) %>% select(MID, Type) %>%
group_by(MID, Type) %>% tally() %>%
arrange(MID, desc(n)) %>% rename(Type_wo_NA = Type)
| MID | Type_wo_NA | n |
|---|---|---|
| 2051R | D | 3 |
| 7004R | A | 3 |
| 7443N | B | 3 |
| 9265T | C | 3 |
Table from Data Excluding NAs
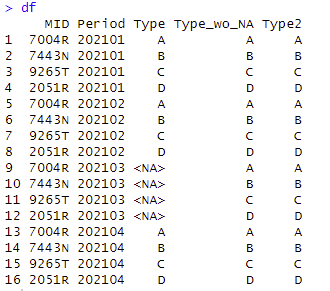
Final result
The previous data table is joined with the original data, adding the ‘Type_wo_NA’ field.
To populate the resulting ‘Type2’ field, the function takes the value from the ‘Type’ field unless it’s an NA error. This evaluation is made possible using the ‘Coalesce’ function from the Dplyr package.
1
2
3
4
5
6
# Left Join Table of Unique Type + Coalesce
df <- df %>% dplyr::left_join(df_noNA[,c(1:2)], by = c('MID'))
df$Type2 <- dplyr::coalesce(df$Type, df$Type_wo_NA)
The final results can be seen in the Type2 field.
__
End of Post