DP19 Classification Categories using Dplyr
Description
Having initial data with some fields about the characteristics of the record the task is to create a special classification field. For that purpose, an evaluation with conditional statements will be performed on each record to give them the final classification.
In this example, it is used the data of interactions with the patients and features about the contact and the outcome. Using the function mutate from the dplyr package helper columns will be added, then group_by to get a recap by categories. This method is similar to the Excel pivot tables.
Link to the Complete Script in Github
R Script - Classification Categories using mutate
First Classification: Patient-Centered Interaction (PCI)
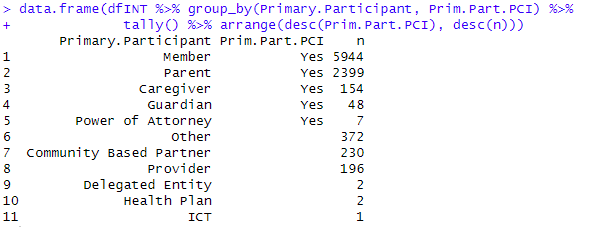
From all the options for Primary.Participant it is required to classify a subgroup as “Patient-Centered Interaction” or PCI. So the field “Prim.Part.PCI” is added as a helper column:
1
2
3
4
5
6
7
# Adding PCI classification
dfINT <- dfINT %>%
dplyr::mutate(Prim.Part.PCI = if_else(Primary.Participant %in% c("Member",
"Parent","Caregiver","Guardian","Power of Attorney"),"Yes", "")) #
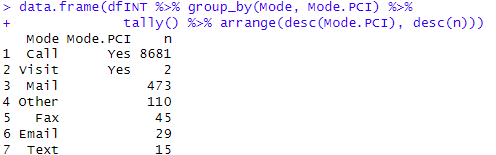
# Recap of fields: Primary.Participant and Prim.Part.PCI
data.frame(dfINT %>% group_by(Primary.Participant, Prim.Part.PCI) %>%
tally() %>% arrange(desc(Prim.Part.PCI), desc(n)))
This recap results shows the Primary Participants that were classified as PCI:
Second Classification: Mode PCI
Similarly, a classification field Mode.PCI is added depending on the Mode of contact:
1
2
3
4
5
6
# Mode.PCI field for interactions with Call, Visit, Videoconference
dfINT <- dfINT %>% mutate(Mode.PCI = if_else(Mode %in%
c("Call", "Visit", "Videoconference"), "Yes", ""))
# Results
data.frame(dfINT %>% group_by(Mode, Mode.PCI) %>%
tally() %>% arrange(desc(Mode.PCI), desc(n)))
Final “Interaction Classification” field
Logic
The next is the logic to get the “Interaction Classification” field, using the previously defined helper fields.
| Final Result | Conditions |
|---|---|
| “Unable to Reach (UTR)” | Outgoing.Contact.Result == “Unable to Reach (UTR)” |
| “Completed Pt. Centered Interaction” | Prim.Part.PCI == “Yes” & Mode.PCI == “Yes” |
| “Other Interactions” | if none of the above conditions were met |
Code
1
2
3
4
5
6
7
8
9
10
11
12
# Int_Classification
# Without Other Completed Int. w/Patient
dfINT <- dfINT %>%
dplyr::mutate(Interaction_Classification =
if_else(Outgoing.Contact.Result == "Unable to Reach (UTR)",
"Unable to Reach (UTR)",
if_else(
Prim.Part.PCI == "Yes" &
Mode.PCI == "Yes",
"Completed Pt. Centered Interaction",
"Other Interactions")))
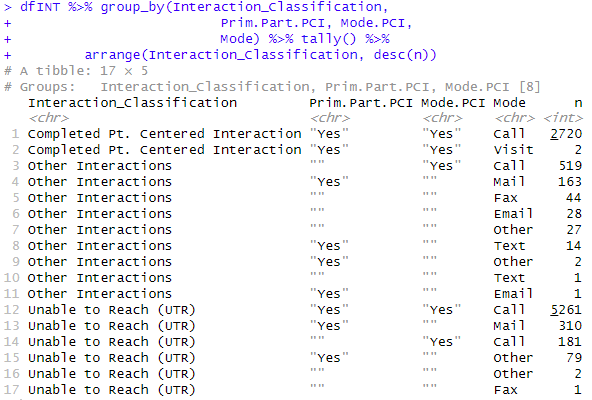
Result of Classification using a Helper column
Summarizing the results:
1
2
3
4
dfINT %>% group_by(Interaction_Classification,
Prim.Part.PCI, Mode.PCI,
Mode) %>% tally() %>%
arrange(Interaction_Classification, desc(n))
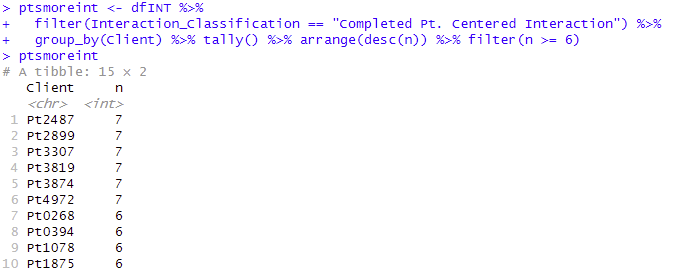
Exploring Specific groups. Data Mining
Using dplyr with specific filters, we can return to the interaction level and examine specific groups, such as patients who received more than 6 interactions.
1
2
3
4
dfINT %>% filter(Interaction_Classification == "Completed Pt. Centered Interaction") %>%
group_by(Client) %>% tally() %>% arrange(desc(n)) %>% filter(n >= 6)
Results in R console
__
End of Post